I have written about chromatography, gas chromatography (GC) and even gas chromatography with mass spectrometry (GC-MS). Here they are (in no particular order):
- GC-MS is not perfect: The case study of methamphetamine
- Why do I see two peaks on my EtOH analysis using HS-GC-FID?
- What is the difference between HS-GC-FID and GC-MS?
- Drugs of Abuse (DOA) analysis performed today in the US
- Fool’s Gold or Real Gold? HS-GC-FID is it specific or selective or neither?
- What is a Gas Chromatography column and why should I care?
- The case for raw data: “Integration” in Gas Chromatography: How to make an innocent person guilty in a DUI case by manipulating the software
- What’s Salt Got to Do With It? Salting Out Effect
- Why forensic science testing for DUI BAC determination is the silly sister of analytical science: Good Laboratory Practice
- A large problem in Gas Chromatography: No uniform standard for GC run position or composition
- Crimping a Headspace Vial in Gas Chromatography: How not putting the “lid on the jar” can lead to disaster in a DUI case
- Gas Chromatography and why is it is so important to Pennsylvania DUI arrests
- Why saying the sample was run on a gas chromatograph is nearly meaningless
- The Carry-over Effect: Lack of Blanks between tests leads to false positive or inflated BAC results
- If he was an analytical chemist or a DUI defense attorney he would say “Show me the separation!!”
- The variables of the Gas Chromatography process
- In analytical chemistry for ETOH and Drugs of Abuse determination, it is very true that the matrix is all around us but truly is ignored
- Personal bias is bad, but analytical bias spells disaster in DUI BAC and Drugs of Abuse testing
- Carryover effect part Deux: Autodilution may be part of the problem for false blood results in DUI
- Internal standard the likely culprit for inaccurate BAC results
- Carryover effect part 3: Flushing of inert gas is not enough to prove there is no carryover
- How do they make the squiggly lines turn into a magic number: Area under the peak
- When tall and skinny is always beautiful? Fundamental principles of gas chromatography
One of the key prerequisites to quantification (which answers the question the question of: How much do we have?) in any form of chromatography is that we must first have unique separation of molecules. Separation first, then quantification. Separation. Separation. Separation. It is very difficult or impossible to reliably and validly quantify mixtures.
But how do we know when or if we have separation? If we do not have separating, then we have co-eluting compounds. But that begs the question doesn’t it? If the two compounds come out at the same time and there is not clear signs such as a doublet or peak-tailing, then how can we know what we see is the product of unique separation, meaning there is no co-elution?
Considering that there are over 56 million organic and inorganic substances and 62 million sequences registered with CAS, co-eultion is a very real theoretical possibility. (CAS REGISTRYSM, the gold standard for substance information, is the only integrated, comprehensive source of chemical information from a full range of disclosed material including patents, journals, and reputable web sources. When you need to positively identify a chemical substance, you can rely on the authoritative source for chemical names and structures of CAS REGISTRY. You can also identify your substance of interest by its CAS Registry Number, which is the best way to identify a substance, regardless of what name you have for it.)
It is a fascinating problem if you think about it.
Wouldn’t it be great if there was software that could take all of the raw data that a GC-MS machine gathers regardless of manufacturer and scrutinize the data to reveal if there is co-elution?
Well, there is.
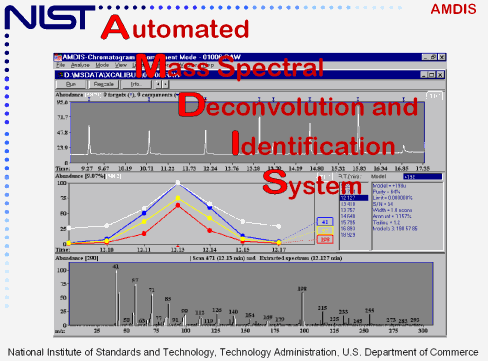
The Automated Mass Spectral Deconvolution and Identification System (AMDIS) is a computer program that extracts spectra for individual components in a GC/MS data file and identifies target compounds by matching these spectra against a reference library. It was developed at NIST with support from the United States Department of Defense and is freely available. Source: http://chemdata.nist.gov/mass-spc/amdis/
AMDIS was developed by our federal government to scrutinize GC-MS data from all software programs. With the support of the the DoD, through the Defense Threat Reduction Agency, AMDIS originally was developed for verification of the Chemical Weapons Convention. It is a very powerful tool that can be used as a “black box” device to identify compounds to try to discount for co-elution. AMDIS is now available to the general analytical chemistry community free of charge.
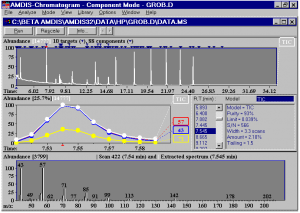
AMDIS takes a chromatogram (either GC/MS or LC/MS) identifies the location of any components, purifies the mass spectra for each component, and then searches a user-specified library for matching compounds.
AMDIS examines spectra by:
- deconvolution
AMDIS does a true deconvolution of the data. Even if there is no available background for subtraction, AMDIS can still extract clean spectra. - treatment of noise
AMDIS does a complete analysis of noise and uses this information for component perception - correction for baseline drift
The chromatogram does not have to have a flat baseline. AMDIS determines the baseline for each component. - extraction of closely co-eluting peaks
AMDIS can extract separate components that peak within a single scan of each other.
Unlike a traditional identification algorithms, AMDIS includes uncertainties in the deconvolution, purity, and retention times in the match factor. The final match factor is a measure of both the quality of the match and of the confidence in the identification.
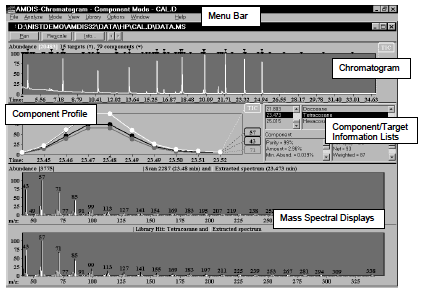
Figure 1: The CONFIRM Window allows you to see the data used in the identification in more detail than in the Results Window. In the Confirm Window, you can view either a single chromatogram or more than one chromatogram.
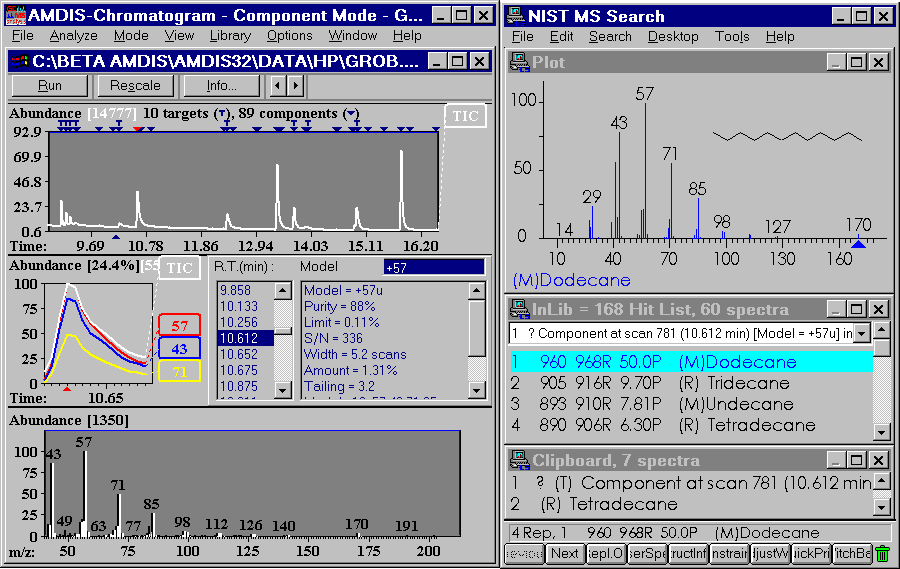
Figure 2: After analyzing a data file in AMDIS you may find that a couple of components have not been identified in the specified library. When you search NIST for individual compounds, you can also use a split screen to view AMDIS and NIST simultaneously.
It exists.
It is free.
So, why doesn’t your forensic science laboratory use it to remove the possibility of error in terms of the reported qualitative and the quantitative result?
Beats me.


